ModelBricks
Complex molecular networks often can be constructed from simpler modules that we call bricks. The adaptive behaviour of living cells is shown to be represented as a combination of functional motifs that reproduce different patterns of cell response to a signal – from simple linear and sigmoidal responses to more complex behaviours like toggle switches and oscillators. The topology of these modules can be captured in graphical form and represented as a wiring diagram in SBGN. However, to understand why these models work the way they do, one must develop a precise mathematical description of molecular circuitry and describe it in a different type of a brick, that we call an executable brick or a ModelBrick.
Process Description ModelBricks
A computable ModelBrick is essentially a small model that is thoroughly annotated, and minted a DOI for a permanent reference. A ModelBrick is a small model that adds a mathematical description and simulation context to an SBGN brick. To enable reproducibility, it is thoroughly annotated, and minted a Digital Object Identifier (DOI) for a permanent reference. Element annotations will include stable identifiers such as PubMed PMIDs for models and parameters, Gene Ontology GO terms for processes, Systems Biology Ontology SBO terms for model elements, Reactome IDs for reactions, ChEBI and UniProt for species, as well as identifiers from other public databases such as KEGG.
The ModelBricks project is led by Michael Blinov (blinov@uchc.edu) and Ann Cowan (acowan@uchc.edu). It is a part of the Virtual Cell modeling and simulation framework (http://vcell.org). VCell BioModels composed of ModelBricks will be interoperable with the SBML Hierarchical Model Composition standard. Different modeling techniques can be used to link multiplex bricks to computable ModelBricks. A ModelBrick comprising a reaction network is a computable extension of of SBGN-PD representation. Kinetic laws and numerical parameters need to be added to such brick in order to make it computable. Rule-based features may be added to identify each species in a model as a composition of uniquely identifiable molecules, and accounts for internal species connectivity and posttranslational modifications.
Tyson and collaborators suggested in their seminal paper “Sniffers, buzzers, toggles and blinkers: dynamics of regulatory and signaling pathways in the cell” (PubMed ID:12648679) that complex molecular networks, like electrical circuits, can be constructed from simpler modules. Below we implement these modules as SBGN bricks and as computable ModelBricks reproducing plots from this paper.
Protein synthesis and degradation
 |
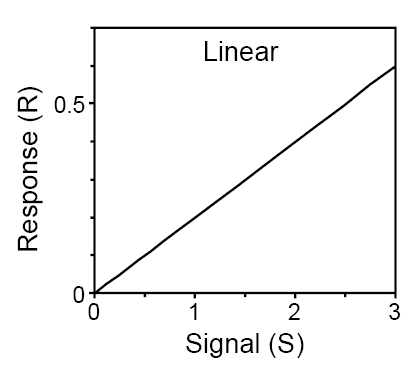 |
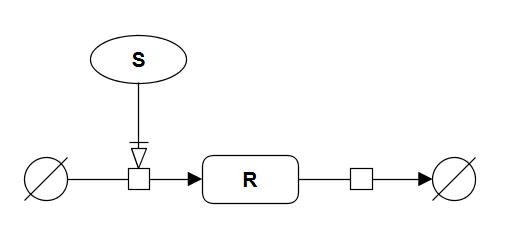
If synthesis of a protein R is regulated by a signal S, with a constant degradation, then R is characterised by a linear response: the amount of R is directly proportional to the strength of signal S. SBGN-PD brick alove provides visualisation of this module. The VCell model Tyson_2003_1a.vcml provides an executable counterpart for this brick.
Protein phosphorylation
 |
 |
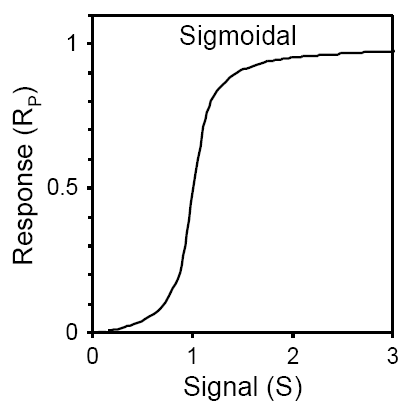 |
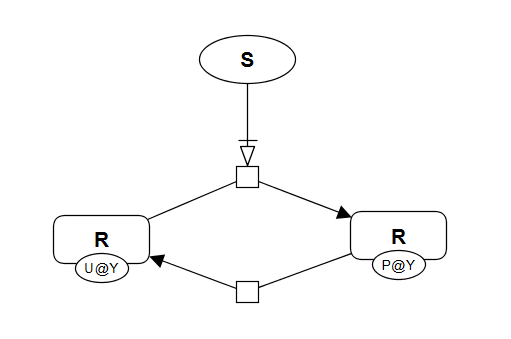
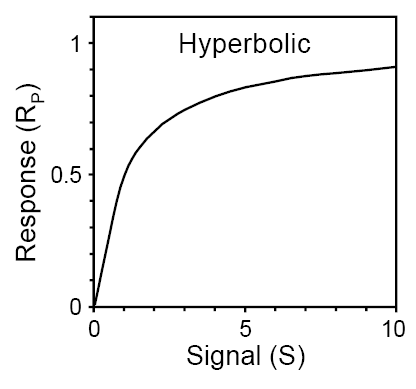
If phosphorylation of a protein R is regulated by a signal S, then the amount of phosphorylated RP is characterised by either a hyperbolic or a sigmoidal response. The visualisation cannot provide the exact response pattern, as it depends on the kinetics of phosphorylation and dephosphorylation reactions. Below is SBGN brick that includes visualisation of this motif. This brick corresponds to two different executable bricks reproducing both signal-response patterns: VCell BioModel Tyson_2003_1b.vcml provides an executable counterpart for this brick for a hyperbolic response, while VCell BioModel Tyson_2003_1c.vcml provides an executable counterpart for a sigmoidal response.
SBGN-PD brick ModelBrick (VCell) Hyperbolic ModelBrick (VCell) Sigmoidal ModelBrick (SBML) Hyperbolic ModelBrick (SBML) Sigmoidal
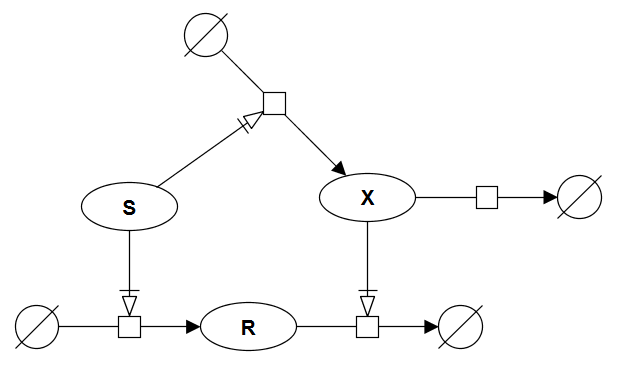
Perfectly adapted signal-response
 |
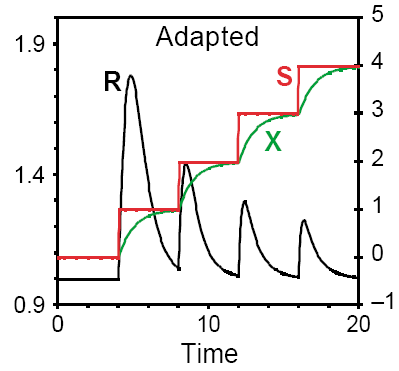 |
Although the signalling pathway exhibits a transient response to changes in signal strength, its steady-state response R is independent of S. Such behaviour is typical of chemotactic systems, which respond to an abrupt change in attractants or repellents, but then adapt to a constant level of the signal. The human sense of smell operates the same way. Below is SBGN brick that provides visualisation of this motif. The VCell BioModel Tyson_2003_1d.vcml provides an executable counterpart for this brick.